Biotechnology news
Researchers make DNA storage a reality
Researchers at the EMBL-European Bioinformatics Institute (EMBL-EBI) have created a way to store data in the form of DNA – a material that lasts for tens of thousands of years. The new method, published today in the journal Nature, makes it possible to store at least 100 million hours of high-definition video in about a cup of DNA. Ads by Google DNA Methylation Products - MethylEasy™ DNA Bisulfite Kit reduces DNA loss by over 90%. - www.geneticsignatures.com There is a lot of digital information in the world – about three zettabytes' worth (that's 3000 billion billion bytes) – and the constant influx of new digital content poses a real challenge for archivists. Hard disks are expensive and require a constant supply of electricity, while even the best 'no-power' archiving materials such as magnetic tape degrade within a decade. This is a growing problem in the life sciences, where massive volumes of data – including DNA sequences – make up the fabric of the scientific record. "We already know that DNA is a robust way to store information because we can extract it from bones of woolly mammoths, which date back tens of thousands of years, and make sense of it," explains Nick Goldman of EMBL-EBI. "It's also incredibly small, dense and does not need any power for storage, so shipping and keeping it is easy." Reading DNA is fairly straightforward, but writing it has until now been a major hurdle to making DNA storage a reality.
There are two challenges: first, using current methods it is only possible to manufacture DNA in short strings. Secondly, both writing and reading DNA are prone to errors, particularly when the same DNA letter is repeated. Nick Goldman and co-author Ewan Birney, Associate Director of EMBL-EBI, set out to create a code that overcomes both problems. "We knew we needed to make a code using only short strings of DNA, and to do it in such a way that creating a run of the same letter would be impossible. So we figured, let's break up the code into lots of overlapping fragments going in both directions, with indexing information showing where each fragment belongs in the overall code, and make a coding scheme that doesn't allow repeats. That way, you would have to have the same error on four different fragments for it to fail – and that would be very rare," says Ewan Birney.
The new method requires synthesising DNA from the encoded information: enter Agilent Technologies, Inc, a California-based company that volunteered its services. Ewan Birney and Nick Goldman sent them encoded versions of: an .mp3 of Martin Luther King's speech, "I Have a Dream"; a .jpg photo of EMBL-EBI; a .pdf of Watson and Crick's seminal paper, "Molecular structure of nucleic acids"; a .txt file of all of Shakespeare's sonnets; and a file that describes the encoding. "We downloaded the files from the Web and used them to synthesise hundreds of thousands of pieces of DNA – the result looks like a tiny piece of dust," explains Emily Leproust of Agilent. Agilent mailed the sample to EMBL-EBI, where the researchers were able to sequence the DNA and decode the files without errors. "We've created a code that's error tolerant using a molecular form we know will last in the right conditions for 10 000 years, or possibly longer," says Nick Goldman. "As long as someone knows what the code is, you will be able to read it back if you have a machine that can read DNA." Although there are many practical aspects to solve, the inherent density and longevity of DNA makes it an attractive storage medium. The next step for the researchers is to perfect the coding scheme and explore practical aspects, paving the way for a commercially viable DNA storage model.
Type II Diabetes and Alzheimer's Connection
A research team in Israel has devised a novel approach to identifying the molecular basis for designing a drug that might one day decrease the risk diabetes patients face of developing Alzheimer's disease.
A recent study suggests that people who suffer from type 2 diabetes face twice the risk of developing Alzheimer's disease later in life compared to those who do not have diabetes. The link these diseases share relates to the formation of two types of peptide deposits that aggregate, or clump together. Peptides are chains of amino acids; longer chains form proteins. One type of peptide, called amyloid beta, is found in Alzheimer plaques in neurons of the brain. The other type, amylin, is found in the pancreas and the brain. Two years ago, researchers found both molecules in the pancreas of diabetic patients, and in both diseases their presence has been linked to the progression of the disease state. To explore the hypothesis that interactions between the two molecules might play a critical role in the self-assembly of peptides that leads to protein aggregation, Yifat Miller, assistant professor from Ben-Gurion University of the Negev, Beer-Sheva, Israel, characterized the way the two protein molecules interact with each other through an examination of their structure. It was the first analysis of its kind. "By identifying the specific 'hot regions' of these peptides that strongly interact with each other, our study may provide insight into the link between type 2 diabetes and Alzheimer disease," Miller says. "We believe that preventing these interactions by developing a drug will decrease the risk that type 2 diabetes patients face of developing Alzheimer's disease later life."
Caught in the Act: Researchers Capture Key Moments in Cell Death
Scientists at the Walter and Eliza Hall Institute have for the first time visualised the molecular changes in a critical cell death protein that force cells to die.
Using the Australian Synchrotron, Dr Czabotar and colleagues were able to obtain detailed three-dimensional images of Bax changing shape as it moved from its inactive to active form. The active form ruptures mitochondrial membranes, removing the cell's energy supply and causing cell death. "By using the powerful X-ray beams created by the synchrotron, we obtained structures of Bax that were really exciting," Dr Czabotar said. "Bax is activated when small protein fragments called BH3-peptides bind to it. We saw that these peptides open up the Bax molecule like a key unlocking a padlock. This unlocked form of Bax can bind to another Bax molecule, which can then form larger Bax complexes that can go on to break up membranes in the cell.
"As well as explaining the detail of how cell death occurs, our research could provide clues about how to design potential new therapeutic agents that target Bax," Dr Czabotar said. "Now that we can see how Bax changes its shape to move from the inactive to the active form, it may be possible to block Bax activation, to prevent cell death in conditions such as neurodegenerative disorders, where illness is caused by excessive cell death. Similarly, agents that drive Bax into its active form could force immortal cells such as cancer cells to die, providing the basis for a potential new class of anti-cancer agents."
Visualization of molecular changes in a critical cell death protein
that force cells to die.
(Credit: Image courtesy of Walter and Eliza Hall Institute)
New DNA Vaccine. A Dramatic Step Forward
New and increasingly sophisticated vaccines are taking aim at a broad range of disease-causing pathogens, targeting them with greater effectiveness, at lower cost and with improved measures to ensure safety. To advance this quest, a research team led by Roy Curtiss, director of the Center for Infectious Diseases and Vaccinology, and Wei Kong, a research assistant professor, at Arizona State University's Biodesign Institute have taken a dramatic step forward, revealing the design of a universal platform for delivering highly potent DNA vaccines, by employing a cleverly re-engineered bacterium to speed delivery to host cells in the vaccine recipient. "The technology that we're describing in this paper can be used to develop a vaccine against any virus, any parasite, any fungus, whereas this was never possible before the development of recombinant attenuated bacterial strains like those produced in our lab," Curtiss says. The experimental vaccine described in the new research demonstrated complete protection from influenza in mice, but Wei Kong, the leading author of the new study stresses that the innovative technique could be applied to the rapid manufacture of effective vaccines against virtually any infectious invader at dramatically reduced cost and without risk to either those vaccinated or the wider public. "By delivering the DNA vaccine using a recombinant attenuated bacterium, we can get 10,000-100,000 doses per liter of culture," Kong says, an improvement of 3-4 orders of magnitude over use of the naked plasmid DNA, which must be painstakingly isolated from bacteria before injection.
The group's research results appear in the online Early Edition (EE) of the Proceedings of the National Academy of Sciences, the week of November 5, 2012. Designing a vaccine that is both safe and effective presents a kind of Catch-22 for researchers. Live pathogenic strains typically generate a robust immune response, mimicking natural infection, but many challenges exist in terms of ensuring such strains do not cause illness or escape into the environment, where they have the potential to remain viable. Killed pathogen strains or vaccines produced from pathogen subunits sacrifice some of their immunogenic effectiveness for enhanced safety, and may require subsequent booster doses to ensure continued effectiveness. The Curtiss team has worked to combine safety and effectiveness in orally administered vaccines that can be produced at a fraction of the cost of traditional methods. To do this, they have pioneered techniques using Salmonella — the notorious agent associated with food-borne illness — as a cargo vessel to deliver a suite of disease antigens to the recipient. The result has been the development and ongoing refinement of so-called RASVs (for recombinant attenuated Salmonella vaccines), capable of provoking an intense, system-wide immune response and conferring effective immunity. One of the key innovations developed earlier by Wei Kong and other members of the Curtiss group, is a specialized Salmonella strain that can be timed to self-destruct in the body once it has carried out its immunization duties. To create this strain, the researchers modified the bacterium in such a way that it can only survive on a non-naturally occurring form of sugar. Once the Salmonella cells exhaust their store of specialized sugar, supplied to them as part of the vaccine, they are unable to maintain the integrity of their cell walls and they essentially implode. "This crucial safety feature ensures that Salmonella are unable to persist as living organisms to survive if excreted into the environment," says Kong.
This self-destruct feature can be fine-tuned so that the bacteria fully colonize host cells, provoking a strong response from both humoral and cell-mediated arms of the immune system. Inside host tissues, recombinant Salmonella are able to synthesize protective antigens, releasing their contents when they become unstable and lyse into the cytosol. The group demonstrated the effectiveness of these delayed-lysis bacteria in vaccine experiments with a variety of pathogens, including influenza and mycobacteria (causative agent of tuberculosis) and an RASV vaccine developed in the Curtiss lab against infant pneumonia is currently in FDA Phase I clinical trials. This earlier work focused on producing protective protein antigens in a bacterium, which would subsequently release a dose of these antigens when the bacterial cell lysed within host cells and tissues. In the latest research, the group sought to turn a delayed-lysis Salmonella strain into a universal DNA vaccine delivery vehicle. DNA vaccines stimulate cellular and humoral immune responses to protein antigens through the direct introduction of genetic material, prompting host cells to manufacture specific gene products. This is a crucial advance as it allows for the production of antigens that undergo host cell modification through the addition carbohydrates — a process known as glycosylation. Such modified antigens, which occur in a broad range of pathogenic viruses, fungi and parasites require synthesis by host cells, rather than by the attenuated bacteria. "Here, we were able to deliver a vaccine whose DNA sequence induces the immunized individual to make the protective glycoprotein the way you would during a viral infection," Curtiss says. Previous efforts to achieve this advance for delivery of DNA vaccines by bacteria date to 1995, but only now has such work come to fruition. A number of key modifications to the delayed-lysis RASV were required for this feat, and the Kong and Curtiss team has worked intensively over the past 5 years to achieve them. A hyperinvasive form of Salmonella was constructed through recombinant DNA methods in order to maximize the vaccine vector's ability to invade host cells and become internalized. Following host cell uptake, Salmonella are encased in a membrane-bound endosome known as the Salmonella Containing Vacuole. The RASV was further modified to permit escape from the endosome so that the mature bacterium could spew its immunogenic contents into the host cell's cytosol. Finally, further revisions to the Salmonella strain were applied to diminish the pathogen's ability to cause host cell death, which would prevent the DNA vaccine from migrating to the host cell nucleus to induce the synthesis of protective antigens necessary for the immune response.
The authors note that their orally-administered RASV is markedly superior to earlier efforts which introduced DNA vaccines by means of intramuscular injection or gene gun. These methods fail to deliver the vaccine to both mucosal tissues and certain internal lymphoid tissues, vital to a sustained, protective immunity. "We can protect mice to doses of influenza that would be lethal were they not effectively immunized," Curtiss says, adding that "RASV safety has been established in mice just two hours old as well as in pregnant and immunodeficient mice." Influenza spreads around the world in seasonal epidemics, resulting in about three to five million yearly cases of severe illness and about 250,000 to 500,000 yearly deaths, rising to millions in some pandemic years. Current manufacture of influenza vaccines requires use of chick embryos or cell culture methods. Global capacity is limited, making sufficient vaccine to immunize everyone impossible. Adding to concerns about managing future naturally occurring influenza epidemics is the potential for bioterrorists to produce weaponized influenza strains created using plasmid-based reverse genetics systems. "Increasing the speed of producing a matching vaccine is key in the context of response to an influenza epidemic," Kong says. (Macroevolution Net)
A Mammalian lung, 3D. Researchers create model of mysterious region
Oct. 4, 2012
Within the extraordinarily dense network of pathways in a mammalian lung is a common destination. There, any road leads to a cul-de-sac of sorts called the pulmonary acinus. This place looks like a bunch of grapes attached to a stem (acinus means "berry" in Latin). Scientists have struggled to understand more specifically what happens in this microscopic, labyrinthine intersection of alleys and dead ends. To find out, a research team led by the University of Iowa created the most detailed, three-dimensional rendering of the pulmonary acinus. The computerized model, derived from mice, faithfully mimics each twist and turn in this region, including the length, direction and angles of the respiratory branches that lead to the all-important air sacs called alveoli. "The imaging and image analysis methods described here provide for branch morphometry at the acinar level that has not been available previously," the researchers write in the paper, published this week in the online early edition of the Proceedings of the National Academy of Sciences. The model is important, because it can help scientists understand where and how lung diseases emerge as well as the role the pulmonary acinus plays in the delivery of drugs, such as those commonly administered with inhalers."These methods allow us to understand where in the lung periphery disease begins and how it progresses," says Eric Hoffman, professor in the departments of radiology, medicine, and biomedical engineering at the UI and corresponding author on the paper. "How do gases and inhaled substances get there and do they accumulate in one or another acinus? How do they swirl around and clear out? We just don't have a complete understanding how that happens. "As an example, Hoffman said the model could be used to determine how smoking-induced emphysema originates. "It has been hypothesized recently that it begins with the loss of peripheral airways rather than the lung air sacs," he says, citing ongoing research by James Hogg at the University of British Columbia, who was not involved in this study. It also could shed light and lead to more effective treatment of chronic obstructive pulmonary disease, which causes irreversible damage to the lung, says Dragos Vasilescu, first author on the paper who based his thesis on the research while a graduate student at the UI.
For years, the best that lung anatomy pioneers such as study co-corresponding author Ewald Weibel, professor emeritus of anatomy at the University of Bern, could do to study specific areas of a lung was to make measurements in two dimensions or create 3D casts of a lung's air spaces. The techniques, while giving the earliest insights into a lungs's makeup and functioning, had their limitations. For one, they did not directly replicate a lung's structure in real life, and they could not convey how various parts act together as a whole. Yet advances in imaging and computation have enabled researchers to more fully explore how gases and other inhaled substances act in the lung's furthest recesses. In this study, the team worked with 22 pulmonary acini culled from young and old mice. They then set to "reconstruct" the acini based on micro computed tomography imaging of scanned lungs in mice and extracted from them. The extracted lungs were preserved in a way that kept the anatomy intact—including the tiny air spaces required for successful imaging. From that, the researchers were able to measure an acinus, estimate the number of acini for each mouse lung and even count the alveoli and measure their surface area.
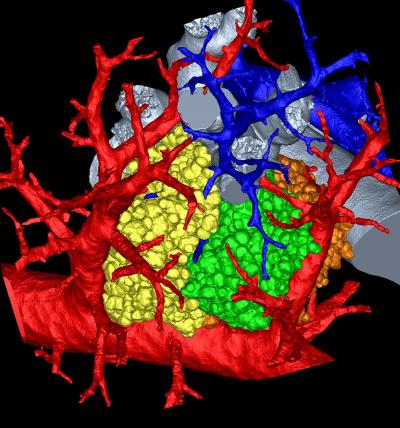
|
The image pictured here shows a mouse's pulmonary acinus, the terminals where gases and blood mix in a lung and whose function remains a mystery. Image: Dragos Vasilescu, University of Iowa and the University of British Columbia. |
The mouse lung, in its structure and function, is remarkably similar to the human lung. That means researchers can alter the genetics of a mouse and see how those changes affect the peripheral structure of the lung and its performance. Already, the researchers found in the current study that mouse alveoli increase in number long past the two weeks that at least one previous study had indicated. Hoffman adds that a separate study is needed to determine whether humans, too, increase the number of air sacs past a certain, predetermined age. The researchers next aim to use the model to more fully understand how gases interact with the bloodstream within the acini and the alveoli. "Our imaging and image-analysis methodologies enable new ways to investigate the lung's structure and can now be used to further investigate the normal healthy-lung anatomy in humans and be used to visualize and assess the pathological changes in animal models of specific structural diseases," says Vasilescu, who is a postdoctoral researchfellow at the University of British Columbia (Macroevolution Net).
Contact
Radmile Matejčić 2
51000 Rijeka
Croatia
Tel. +385 51 584 550
ured@biotech.uniri.hr
OIB (VAT): 64218323816
IBAN: HR3724020061400006958
Radmile Matejčić 2, O-047
http://studentska.uniri.hr/
Biotechnology for the Life Sciences e-mail :














